R Workshop: Module 3 (1)
Bobae Kang
March 28, 2018
This page contains the notes for the first part of R Workshop Module 3: Data Analysis with R, which is part of the R Workshop series prepared by ICJIA Research Analyst Bobae Kang to enable and encourage ICJIA researchers to take advantage of R, a statistical programming language that is one of the most powerful modern research tools.
Links
Click here to go to the workshop home page.
Click here to go to the workshop Modules page.
Click here to view the accompanying slides for Module 3, Part 1.
Navigate to the other workshop materials:
Data Analysis with R (1): Getting started with tidyverse

Source: tidyverse.org
In Module 3, we will be exploring several tidyverse packages that offer powerful functionalities for data analysis.
Getting Ready
Installing the packages
tidyverse is a set of R packages as well as an opinionated philosophy underlying these packages on how the practice of working with data in R should be. Although it is not the only way of working with data in R, tidyverse packages have become highly popular among R users.
Installing all tidyverse packages can be easily done with the follwoing command:
# Install from CRAN
install.packages("tidyverse")
# Or the development version from GitHub
# install.packages("devtools")
devtools::install_github("hadley/tidyverse")Installing tidyverse package installs the following:
- core tidyverse packages
ggplot2,dplyr,tidyr,readr,purrr,tibble
- packages to work with specific vector types
hms,stringr,lubridate,forcats
- packages to import data
feather,haven,httr,jsonlite,readxl,rvest,xml2
- packages to facilitate statistical modeling
modelr,broom
Alternatively, each pacakge can be installed separately:
# Install ggplot2
install.packages("ggplot2")
# Install both dplyr and tidyr with a single commend
# with a character vector of the package names
install.packages(c("dplyr", "tidyr"))Importing the packages
Once installed, we can now import the packages using library():
# This imports the core tidyverse packages
library(tidyverse)
# Or import packages separately
library(dplyr)
library(tidyr)In the following, we will explore two powerful tidyverse packages for wrangling tabluar data, dplyr and tidyr.
Manipulating Your Data

Source: tidyverse.org
Key dplyr functions
In this section, we will learn the following dplyr functions:
arange()select()rename()filter()mutate()transmute()left_join()summarise()group_by()ungroup()%>%, the pipe operator
Please note that this section is not intended to be an exhaustive documentation and description of the listed functions–or dplyr package for that matter. For more information, please check out the reference materials listed below.
Sort rows by variables
arrange(tbl, ...)arrange() is used to sort the rows by a selection of column variables. The first argument of arrange() is a tabular data object. In fact, as we will see soon, almost all other functions of tidyverse packages take a data object as the first input. This tabular data can be of the data.frame class or any of its extensions, such as tidyverse’s very own tibble class.
arrange() takes columns by which to sort the data as additional arguments. If multiple columns are provided, the sorting taks place in a hierarchical manner: the data is first sorted by the first column, then by the second column, and so on. By default, sorting is in an ascending order (smaller to larger). Sorting in a descending order can be done by wrapping the column with desc().
Example
The following example sorts, or arranges, the ispcrime data by the county column. To make it manageable, we also use the head() to print only the first few rows:
# without using pipe
head(arrange(ispcrime, county))
# with using pipe
ispcrime %>%
arrange(county) %>%
head()## year county violentCrime murder rape robbery aggAssault propertyCrime
## 1 2011 Adams 218 0 37 15 166 1555
## 2 2012 Adams 205 0 28 16 161 1587
## 3 2013 Adams 222 1 33 21 167 1564
## 4 2014 Adams 222 2 34 13 173 1550
## 5 2015 Adams 227 5 39 9 174 1268
## 6 2011 Alexander 119 0 14 4 101 290
## burglary larcenyTft MVTft arson
## 1 272 1241 36 6
## 2 287 1261 30 9
## 3 297 1204 55 8
## 4 305 1201 39 5
## 5 274 944 43 7
## 6 92 183 11 4As shown above, there are two ways to work with dplyr functions:
- Traditional syntax for using R functions
- Using pipe operators,
%>%
We will take a closer look at %>% below. In the meantime, it is sufficient to note that the object before %>% is fed, or “piped,” into the following function as the input for its first argument.
The folloiwng code illustrates using desc() to sort in a descending order:
# sort by county, in a DESCENDING order
ispcrime %>%
arrange(desc(county), desc(year)) %>%
head() ## year county violentCrime murder rape robbery aggAssault propertyCrime
## 1 2015 Woodford 29 0 5 0 24 227
## 2 2014 Woodford 25 0 4 0 21 283
## 3 2013 Woodford 17 0 10 2 5 349
## 4 2012 Woodford 23 0 3 1 19 405
## 5 2011 Woodford 15 1 4 0 10 350
## 6 2015 Winnebago 2692 22 197 563 1910 8279
## burglary larcenyTft MVTft arson
## 1 35 186 6 0
## 2 50 218 13 2
## 3 81 257 11 0
## 4 65 329 10 1
## 5 90 247 10 3
## 6 1958 5635 618 68Filter rows with conditions
filter(tbl, ...)filter() is used to filter data to keep rows that match given conditions. As in arrange(), the first argument is a tabular data object. Conditional expressions to filter data, which should evaluate as TRUE or FALSE for each row, are given as additional arguments.
Example
The following code filters ispcrime to return a subset of data where year value matches 2015 AND murder value is greater than 0. As before, we use head() to show only the first few rows.
ispcrime %>%
filter(
year == 2015,
murder > 0
) %>%
head()## year county violentCrime murder rape robbery aggAssault propertyCrime
## 1 2015 Adams 227 5 39 9 174 1268
## 2 2015 Bond 5 1 0 0 4 130
## 3 2015 Bureau 53 1 17 4 31 360
## 4 2015 Champaign 918 7 127 205 579 5568
## 5 2015 Coles 173 1 33 14 125 632
## 6 2015 Cook 28791 534 1911 11217 15129 124784
## burglary larcenyTft MVTft arson
## 1 274 944 43 7
## 2 39 84 7 0
## 3 110 244 6 0
## 4 1100 4235 196 37
## 5 146 463 13 10
## 6 20550 90675 12547 1012Select and/or rename variables
select(tbl, ...)
rename(tbl, ...)select() and rename() both take a tabular data object as the first argument input. The following arguments are columns to select or rename. These two functions are quite similar to each other, except that select() returns the selected columns only while rename() keeps all columns.
It is actually possible to renaming each selected column with select() as well. select() can be also used to exclude a section of columns using a minus (-) sign while keeping the rest.
Example
Let’s take a look at an example on how to use select() to select columns, with changing their names in certain cases. The following code select four columns of ispcrime while changing the names of violentCrime and propertyCrime columns.
# select with renaming columns
ispcrime %>%
select(year, county, v_crime = violentCrime, p_crime = propertyCrime) %>%
head()## year county v_crime p_crime
## 1 2011 Adams 218 1555
## 2 2011 Alexander 119 290
## 3 2011 Bond 6 211
## 4 2011 Boone 59 733
## 5 2011 Brown 7 38
## 6 2011 Bureau 42 505As noted earlier, select() can be used to exclude certain columns. The following code selects all columns of ispcrime except violentCrime and propertyCrime:
# excluding columns
ispcrime %>%
select(-violentCrime, -propertyCrime) %>%
head()## year county murder rape robbery aggAssault burglary larcenyTft MVTft
## 1 2011 Adams 0 37 15 166 272 1241 36
## 2 2011 Alexander 0 14 4 101 92 183 11
## 3 2011 Bond 1 0 0 5 58 147 5
## 4 2011 Boone 0 24 8 27 152 563 14
## 5 2011 Brown 0 1 0 6 14 22 1
## 6 2011 Bureau 0 4 3 35 90 405 8
## arson
## 1 6
## 2 4
## 3 1
## 4 4
## 5 1
## 6 2Transform and add variables
transmute(tbl, ...)
mutate(tbl, ...)transmute() and mutate() both take a tabular data object as the first argument input. The following arguments are expressions to transform existing columns or add new ones. An exisiting column is modified with an expression with the same column name. On the other hand, a new column is created with an expression having a new column name.
As with select() and rename(), the two functions here are quite similar to each other, except that transmute() returns the trasnformed/aded columns only while mutate() keeps all columns.
Example
The following code adds a new column named totalCrime, whose value equals the sum of violentCrime and propertyCrime values:
ispcrime %>%
mutate(totalCrime = violentCrime + propertyCrime) %>%
head()## year county violentCrime murder rape robbery aggAssault propertyCrime
## 1 2011 Adams 218 0 37 15 166 1555
## 2 2011 Alexander 119 0 14 4 101 290
## 3 2011 Bond 6 1 0 0 5 211
## 4 2011 Boone 59 0 24 8 27 733
## 5 2011 Brown 7 0 1 0 6 38
## 6 2011 Bureau 42 0 4 3 35 505
## burglary larcenyTft MVTft arson totalCrime
## 1 272 1241 36 6 1773
## 2 92 183 11 4 409
## 3 58 147 5 1 217
## 4 152 563 14 4 792
## 5 14 22 1 1 45
## 6 90 405 8 2 547If it were with transmute(), the result would include only the totalCrime column.
Merge tables
left_join(tbl1, tbl2, by = NULL, ...)left_join() takes two tabular data objects to join as the first two arguments. by takes a chracter vector containing a selection of variables to join tables by. By default, all columns with common names are used. The ouput is a merged table where the columns in the second table is added to the first table as new columns where the values of by columns match.
left_join() is named so because it keeps all observations in the first (left) table. There, in fact, are other types of joins including: inner_join(), right_join(), full_join(), semi_join(), and anti_join().
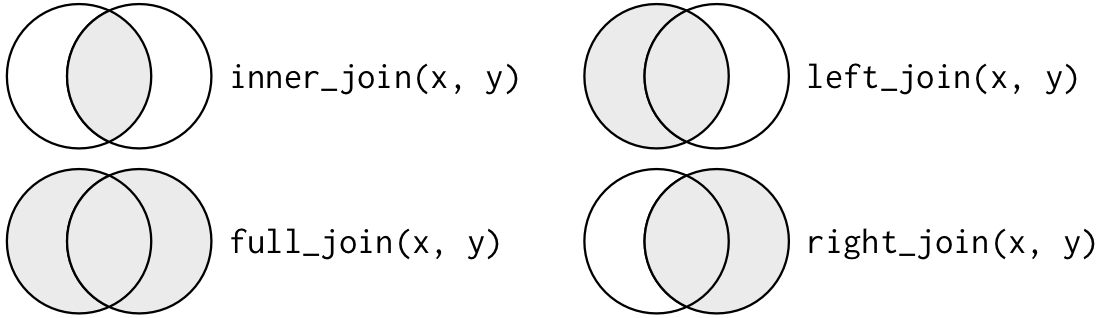
Source: Wickham, H. (2017). R for Data Science
Example
The following code left-joins regions table to ispcrime table on the county values. The regions table has two columns: region and county. Accordingly, the final output is basically the ispcrime table with one extra column, region, that corresponds to the county value:
ispcrime %>%
left_join(regions, by = "county") %>%
head()## year county violentCrime murder rape robbery aggAssault propertyCrime
## 1 2011 Adams 218 0 37 15 166 1555
## 2 2011 Alexander 119 0 14 4 101 290
## 3 2011 Bond 6 1 0 0 5 211
## 4 2011 Boone 59 0 24 8 27 733
## 5 2011 Brown 7 0 1 0 6 38
## 6 2011 Bureau 42 0 4 3 35 505
## burglary larcenyTft MVTft arson region
## 1 272 1241 36 6 Central
## 2 92 183 11 4 Southern
## 3 58 147 5 1 Southern
## 4 152 563 14 4 Northern
## 5 14 22 1 1 Central
## 6 90 405 8 2 CentralAggregate and summarise rows
summarise(tbl, ...)
summarize(tbl, ...)summarise() and summairze() are identical except who they are spelled. This is because the original package author, Hadly Wickham, is an Australian and wrote the names accordingly. Later, due to the demand from American programmers, summarize() was added.
summarise() takes a tabular data object as its first argument, and the following arguments are expressions to “summarize” data. Each of these expressions creates a summary column, which can be given a name just as in the case of transmute() or mutate().
Example
Here, we use summarise() to get the average count of violentCrime and propertyCrime, which are named violentCrimeAverage and propertyCrimeAverage respectively:
ispcrime %>%
summarise(
violentCrimeAverage = mean(violentCrime, na.rm = TRUE),
propertyCrimeAverage = mean(propertyCrime, na.rm = TRUE)
)## violentCrimeAverage propertyCrimeAverage
## 1 500.9702 2912.905Group by variables
group_by(tbl, ...)
ungroup(tbl, ...)group_by is a powerful function that allows us to manipulate a data object in “groups,” which is defined by the discrete values in a column, or a selection of columns. As in other dplyr functions, the first argument is a tabular object. The following arguments are columns by which to group the data.
group_by() and ungroup() are the opposite operations. group_by() creates a data object with groups. ungroup() removes the groups.
Example
The following example shows how a groupd data table looks like:
ispcrime %>%
group_by(year)## # A tibble: 510 x 12
## # Groups: year [5]
## year county violentCrime murder rape robbery aggAssault propertyCrime
## <int> <fct> <int> <int> <int> <int> <int> <int>
## 1 2011 Adams 218 0 37 15 166 1555
## 2 2011 Alexa~ 119 0 14 4 101 290
## 3 2011 Bond 6 1 0 0 5 211
## 4 2011 Boone 59 0 24 8 27 733
## 5 2011 Brown 7 0 1 0 6 38
## 6 2011 Bureau 42 0 4 3 35 505
## 7 2011 Calho~ 13 0 0 0 13 56
## 8 2011 Carro~ 8 0 1 0 7 206
## 9 2011 Cass 12 0 1 0 11 119
## 10 2011 Champ~ 1210 5 127 208 870 5332
## # ... with 500 more rows, and 4 more variables: burglary <int>,
## # larcenyTft <int>, MVTft <int>, arson <int>Now, let’s see how group_by() can be used in combination with another dplyr function. In the following, we group our data by year values and get summaries (average crime counts) for each year:
ispcrime %>%
group_by(year) %>%
summarise(
violentCrimeAverage = mean(violentCrime, na.rm = TRUE),
propertyCrimeAverage = mean(propertyCrime, na.rm = TRUE)
)## # A tibble: 5 x 3
## year violentCrimeAverage propertyCrimeAverage
## <int> <dbl> <dbl>
## 1 2011 538. 3329.
## 2 2012 520. 3192.
## 3 2013 503. 2940.
## 4 2014 464. 2609.
## 5 2015 478. 2481.Chain operations
We have been using the pipe operator, %>%, in our code examples. Now is a good time to take a closer look at it.
data %>% function1(arg2, ...) %>% ...%>% is a binary operator that requires two objects for it to work on. Think of + as another example of a binary operator. In the case of %>% the object preciding the operator has to be the first argument for the function that follows the oprator. In other words, the preceding object is injected into the following function as its first argument. The result of the %>% operator is the output of the function.
In fact, as we have already seen before, multiple functions can be chained, or “piped,” using %>% as long as the output of the first “piping” generates a suitable object for the first argument of the next function to be chained.
with piping, we can have a coding style that is cleaner and more intuitive–that reflects the way we think about how we apply the operations to the object, rather than the tranditional syntax where the function that acts on the last comes at the beginning of the statement:
# piping style
object %>%
function1(arguments1) %>%
function2(arguments2) %>%
function3(arguments3)
# traditional style
function3(
function2(
function1(
object,
arguments1
),
arguments2
),
arguments3
)
# or
function3(function2(function1(object, arguments1), arguments2), arguments3)dplyr in action
Let’s take a look at more concrete examples of using dplyr functions to work on data manipulation and analysis. For those of you who were there at my presentation for the February R&A meeting, these are the some of the examples used in that presentation–now we can understand them better!
Example 1
ispcrime %>%
filter(substr(county, 1, 1) == "D", year %in% c(2014, 2015)) %>%
mutate(totalCrime = violentCrime + propertyCrime) %>%
select(year, county, totalCrime)## year county totalCrime
## 1 2014 De Kalb 2218
## 2 2014 De Witt 182
## 3 2014 Douglas 116
## 4 2014 Du Page 12576
## 5 2015 De Kalb 2173
## 6 2015 De Witt 140
## 7 2015 Douglas 173
## 8 2015 Du Page 12538In this example, we we want to answer the following question: What are the total crime count in 2014 and 2015 for counties whose name starts with letter D?
- We start with
ispcrimedata - We use the pipe operator to feed the
ispcrimedata intofilter()- we have two filtering conditions. First condition is that the first letter of the
countycolumn value matches “D” (substr(county, 1, 1) == "D") - Second condition is that the
yearcolumn value has to be either 2014 or 2015 (year %in% c(2014, 2015))
- we have two filtering conditions. First condition is that the first letter of the
- We use the pipe operator to feed the filtered data into
mutate()- Here we create a new column named
totalCrime, which is the sume ofviolentCrimeandpropertyCrimevalues
- Here we create a new column named
- Finally, we use the filtered and mutated data into
select()- The final output will include
year,county, andtotalCrimecolumns only.
- The final output will include
Example 2
ispcrime %>%
left_join(regions) %>%
group_by(region, county) %>%
summarise(annualAvgCrime = sum(violentCrime, propertyCrime, na.rm = TRUE) / n()) %>%
arrange(desc(annualAvgCrime))## # A tibble: 102 x 3
## # Groups: region [4]
## region county annualAvgCrime
## <fct> <fct> <dbl>
## 1 Cook Cook 182818.
## 2 Northern Du Page 14316.
## 3 Northern Lake 12779.
## 4 Northern Winnebago 12275.
## 5 Northern Will 11078.
## 6 Southern St. Clair 9262.
## 7 Central Sangamon 8876.
## 8 Northern Kane 8332.
## 9 Central Peoria 7229.
## 10 Central Champaign 6567.
## # ... with 92 more rowsIn this example, we we want to answer the following question: what is the annual average of the crime count per county, shown with the region it belongs to and sorted by the average count?
- We start with
ispcrimedata - We use the pipe operator to feed the
ispcrimedata intoleft_join()- This is to add
regioncolumn fromregionstable to our data
- This is to add
- We use the pipe operator to feed the joined data into
group_by()- We group the data by values of the
regionandcountycolumn.
- We group the data by values of the
- We use the pipe operator to feed the grouped data into
summarise()- We summarise data by group to get the annual average crime, a sum of
violentCrimeandpropertyCrime, divided by total counts. Since the input is grouped, the summarising work is also done by group.
- We summarise data by group to get the annual average crime, a sum of
- We use the pipe operator to feed the grouped and summarised data into
arrange()- Here we sort the data by the newly created column,
annualAvgCrime, in a descending order
- Here we sort the data by the newly created column,
More on dplyr
What we have seen is only a tip of an iceberg that is dplyr. Although the aforementioned functions are among the most commonly used in usual data manipulation and analysis tasks, the description and examples here are by no means comprehensive. Also, dplyr offers many other functions to facilitate data manipulation work. I recommend you to check out the following resources to learn more about dplyr:
dplyron tidyverse.orgdplyrCRAN documentationdplyrGithub repository- RStudio. (2017). “Data Manipulation Cheat Sheet”.
Tidying Up Your Data

Source: tidyverse.org
Remember tidy data?

Source: R for Data Science
We have learned the notion of “tidy” data in the second part of the previous module. In short, a dataset is “tidy” when:
- Each column is a variable
- Each row is an observation
- Each cell is a value.
Key tidyr functions
In this section, we will learn the following tidyr functions:
gather()spread()unite()separate()separate_rows()
Please note that, as before, this section is not intended to be an exhaustive documentation and description. For more information, please check out the reference materials listed below.
Make “wide” data longer
gather(tbl, key = "key", value = "value", ..., na.rm = FALSE, ...)gather() is a function to make “wide” data longer, by “gathering” multiple columns into two columns of key-value pairs. The function takes a tabular data object, such as data.frame or tibble, as its first argument. The second argument, key, defines the name of a new column for key and the third argument, value, defines the name of a new column for value. These two arguments have default values that are, not surprisingly, "key" and "value", respectively.
The following arguments (...) are a selection of columns to be used whose name will be made into values for the key column and whose value will be made into the values of the value column.
Make “long” data wider
spread(tbl, key, value, fill = NA, ...)spread() is the opposite of gather(). As in gather(), the first argument is a tabular data object. Then, key is an existing column containing the names of new columns, and value is another column containing the values for the new columns.
Example
Let’s take a look at an example illustrating how gather() and spread() work. First, we prepare a data to be used, which is the familiar ispcrime dataset, except we exclude violentCrime and propertyCrime columns using select(). We assign this to ispcrime_2.
ispcrime_2 <- ispcrime %>%
select(-violentCrime, -propertyCrime) %>%
as_tibble()
ispcrime_2## # A tibble: 510 x 10
## year county murder rape robbery aggAssault burglary larcenyTft MVTft
## <int> <fct> <int> <int> <int> <int> <int> <int> <int>
## 1 2011 Adams 0 37 15 166 272 1241 36
## 2 2011 Alexan~ 0 14 4 101 92 183 11
## 3 2011 Bond 1 0 0 5 58 147 5
## 4 2011 Boone 0 24 8 27 152 563 14
## 5 2011 Brown 0 1 0 6 14 22 1
## 6 2011 Bureau 0 4 3 35 90 405 8
## 7 2011 Calhoun 0 0 0 13 14 41 1
## 8 2011 Carroll 0 1 0 7 38 165 2
## 9 2011 Cass 0 1 0 11 41 71 3
## 10 2011 Champa~ 5 127 208 870 1384 3756 164
## # ... with 500 more rows, and 1 more variable: arson <int>Now, let’s try gather(). We will name the key column type for the type of crime, and the value column count. Then we will “gather” all 8 crime type columns, from murder to arson. The output is a reshaepd dataset that is 8-times “longer” than the input (4,080 = 510 * 8). The name of each input column is now turned into a value for the type column, and the corresponding crime count is a value for the count column.
ispcrime_2 %>%
gather(key = "type", value = "count", murder:arson)## # A tibble: 4,080 x 4
## year county type count
## <int> <fct> <chr> <int>
## 1 2011 Adams murder 0
## 2 2011 Alexander murder 0
## 3 2011 Bond murder 1
## 4 2011 Boone murder 0
## 5 2011 Brown murder 0
## 6 2011 Bureau murder 0
## 7 2011 Calhoun murder 0
## 8 2011 Carroll murder 0
## 9 2011 Cass murder 0
## 10 2011 Champaign murder 5
## # ... with 4,070 more rowsNow, to see that spread() is the opposite process, here we show the result of gather()-ing and than spread()-ing the ispcrime_2 object. The final output is in the same shape as the input.
ispcrime_2 %>%
gather(key = "type", value = "count", murder:arson) %>%
spread(key = type, value = count)## # A tibble: 510 x 10
## year county aggAssault arson burglary larcenyTft murder MVTft rape
## <int> <fct> <int> <int> <int> <int> <int> <int> <int>
## 1 2011 Adams 166 6 272 1241 0 36 37
## 2 2011 Alexander 101 4 92 183 0 11 14
## 3 2011 Bond 5 1 58 147 1 5 0
## 4 2011 Boone 27 4 152 563 0 14 24
## 5 2011 Brown 6 1 14 22 0 1 1
## 6 2011 Bureau 35 2 90 405 0 8 4
## 7 2011 Calhoun 13 0 14 41 0 1 0
## 8 2011 Carroll 7 1 38 165 0 2 1
## 9 2011 Cass 11 4 41 71 0 3 1
## 10 2011 Champaign 870 28 1384 3756 5 164 127
## # ... with 500 more rows, and 1 more variable: robbery <int>Unite multiple columns into one
unite(tbl, col, ..., sep = "_", remove = TRUE)unite() works to combine multiple columns into a single column by concatenating their values. Its first argument is a tabular data object. Then, col is a new column created by “uniting” the following column inputs (...). sep input is a string to be inserted as a seperator between column values when concatenation takes place. The default value for sep is the underscore symbol (_). Finally, remove takes a boolean value for whether removing the original columns or not.
Split a column into many
separate(tbl, col, into, sep = "[^[:alnum:]]+", remove = TRUE, ...)separate() is the opposite of unite(). As in unite(), the first argument is a tabular data object. Here, col is a column to be split into multiple columns. into is a character vector for separated column names, which may have two or more elements. sep is a separator between values, which could be _, or any other. Finally, remove is a boolean for whether removing the original columns or not.
unite() and sperate() are great for cleaning up date or name variables. One common application is to convert two columns for “last name” and “first name” into a single column for “full name”, or vice versa.
Example
The following example illustrates the use of unite() and separate(). We first prepare a data object for this exercise: we left_join() ispcrime and regions and select() only six columns in the following order: year, region, county, volentCrime, and propertyCrime. This transformed data is assigned to ispcrime_3.
ispcrime_3 <- ispcrime %>%
left_join(regions) %>%
select(year, region, county, violentCrime, propertyCrime) %>%
as_tibble()## Joining, by = "county"ispcrime_3## # A tibble: 510 x 5
## year region county violentCrime propertyCrime
## <int> <fct> <fct> <int> <int>
## 1 2011 Central Adams 218 1555
## 2 2011 Southern Alexander 119 290
## 3 2011 Southern Bond 6 211
## 4 2011 Northern Boone 59 733
## 5 2011 Central Brown 7 38
## 6 2011 Central Bureau 42 505
## 7 2011 Southern Calhoun 13 56
## 8 2011 Northern Carroll 8 206
## 9 2011 Central Cass 12 119
## 10 2011 Central Champaign 1210 5332
## # ... with 500 more rowsNow, we will try unite() on region and county columns to create a region_county column. Notice that we do not provide any input for sep argument and, accordingly, the default separator, "_", is used to concatenate the two columns.
ispcrime_3 %>%
unite(col = region_county, region, county)## # A tibble: 510 x 4
## year region_county violentCrime propertyCrime
## <int> <chr> <int> <int>
## 1 2011 Central_Adams 218 1555
## 2 2011 Southern_Alexander 119 290
## 3 2011 Southern_Bond 6 211
## 4 2011 Northern_Boone 59 733
## 5 2011 Central_Brown 7 38
## 6 2011 Central_Bureau 42 505
## 7 2011 Southern_Calhoun 13 56
## 8 2011 Northern_Carroll 8 206
## 9 2011 Central_Cass 12 119
## 10 2011 Central_Champaign 1210 5332
## # ... with 500 more rowsTo see that unite() and separate() are the opposite procedures, we show the result of unite()-ing and separate()-ing the ispcrime_3 data. Note that we have to provide "_" as the sep input to gain back the original input.
ispcrime_3 %>%
unite(col = region_county, region, county) %>%
separate(col = region_county, into = c("region", "county"), sep = "_")## # A tibble: 510 x 5
## year region county violentCrime propertyCrime
## <int> <chr> <chr> <int> <int>
## 1 2011 Central Adams 218 1555
## 2 2011 Southern Alexander 119 290
## 3 2011 Southern Bond 6 211
## 4 2011 Northern Boone 59 733
## 5 2011 Central Brown 7 38
## 6 2011 Central Bureau 42 505
## 7 2011 Southern Calhoun 13 56
## 8 2011 Northern Carroll 8 206
## 9 2011 Central Cass 12 119
## 10 2011 Central Champaign 1210 5332
## # ... with 500 more rowsSplit a row into many
separate_rows(tbl, ..., sep = "[^[:alnum:]]+", ...)We also have separate_rows(), which is similar to separate(), except the former results in a longer table while the latter results in a wider table.
More on tidyr
As in the earlier dplyr case, what we have seen is only part of what tidyr offers. I recommend you to check out the following resources to learn more about tidyr
tidyron tidyverse.orgtidyrCRAN documentationtidyrGithub repository- Page 2 of RStudio. (2017). “Data Import Cheat Sheet”.
References
- Grolemund, G. & Wickham, H. (2017). R for Data Science.
- RStudio. (2017). “Data Import Cheat Sheet”.
- RStudio. (2017). “Data Transformation Cheat Sheet”.
- Tidyverse. (n.d.). dplyr.tidyverse.org.
- Tidyverse. (n.d.). tidyr.tidyverse.org.
- Tidyverse. (n.d.). tidyverse.org.
